
The system architecture is presented here with an apache frontend solution with high availability and scalability: web architecture.
SeqCrawler integrates many open source products. Very extensible by nature, a server can be decomposed in a number of feature blocks.
Each block should be assigned at least 1 cpu (or core) for production systems and 1GB of RAM. Of course, depending on indexation needs (and components), lower or higher configurations will be used.
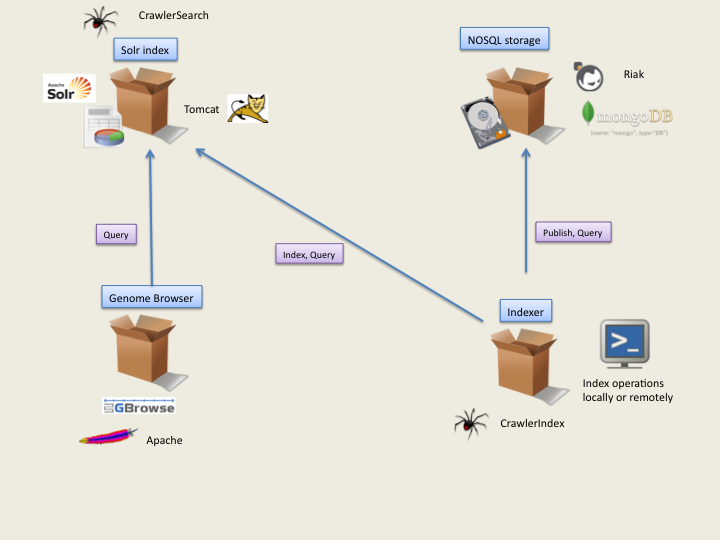
For a look at indexation process, look at CrawlerIndex.
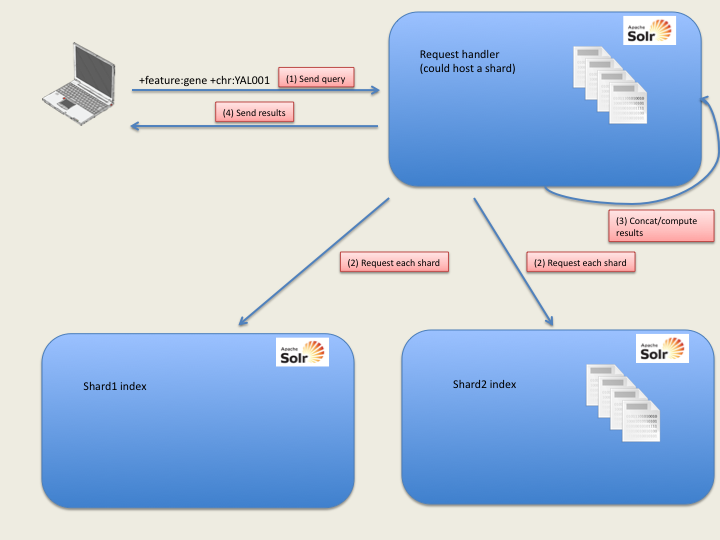
The Solr index supports index sharding e.g. the split of an index in several smaller parts. This is totally transparent in the query. Solr defines some request handlers with default parameters. Though sharding can be specified at request time, we specify it in the configuration file (solrConfig.xml) to ease the query.
If no shard is specified, the Solr instance will at its local index. If shards are set, it will look at the specified instances, which can include itself.
SeqCrawler can store raw sequence data in a NOSQL backend storage (transcript, raw dna) from a GFF/Fasta file. Large data are splitted in lower size documents by the indexer. The structure of a document in the backend is an object accessible with a unique identifier. It is a JSON object:
Example: { "_id" : "BX1234" , "metadata" : " this is a gene" , "shards" : [ "shardid1", "shardid2" ] , "content" : "acgt" }
SeqCrawler provides a basic web interface, dataquery.html, to query the storage.
It can be queried via simple HTTP GET request with parameters:
Dataquery.html will take in charge of assembling the data with internal data shards.
