
CrawlerIndex jar file is in charge of indexing the biological format documents. Many formats are accepted as input, but behaviour won't be the same.
It is based on ReadSeq for many formats, but it implements specific parsers for GFF, EMBL and Fasta formats.
Here is a basic view of the index process:
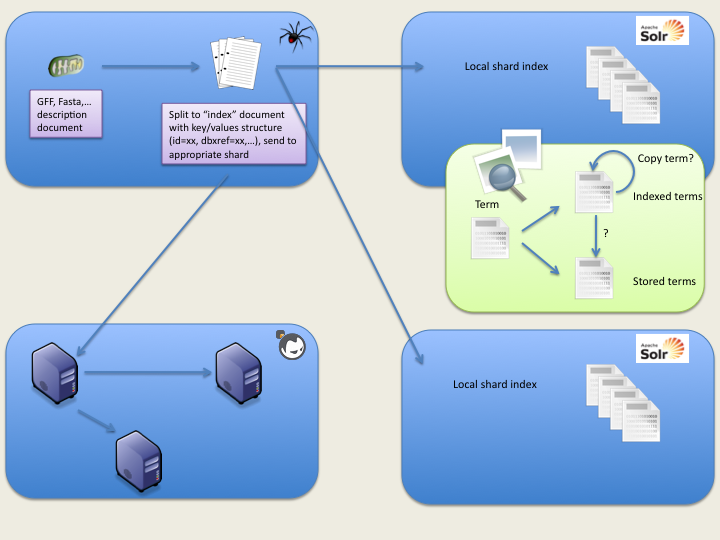
The jar can be executed with: java -jar CrawlerIndex-0.1-jar-with-dependencies.jar The CrawlerIndex contains 3 entry class:
The indexer works with file format handlers. There is a specific handler for several formats (Embl, Fasta, GFF), while the ReadSeq utility is used to handle the other biosequence formats (though readseq handler needs to be specified at command-line).
Storage handlers are handled the same way. Handlers for different backends hide the complexity and dialog with the remote storage. Existing implementations are Riak and MongoDB. A Mock handler is present to simulate a backend for tests, it keeps data in memory. Backends are optional, and only FASTA content is managed with backends in this program.
Logs are managed by the Logback tool, over SLF4J. A basic logger reports to the console with the INFO or above logs. To modify the logger, a logback.groovy can be created in classpath. To do so, refer to the logback tutorial.